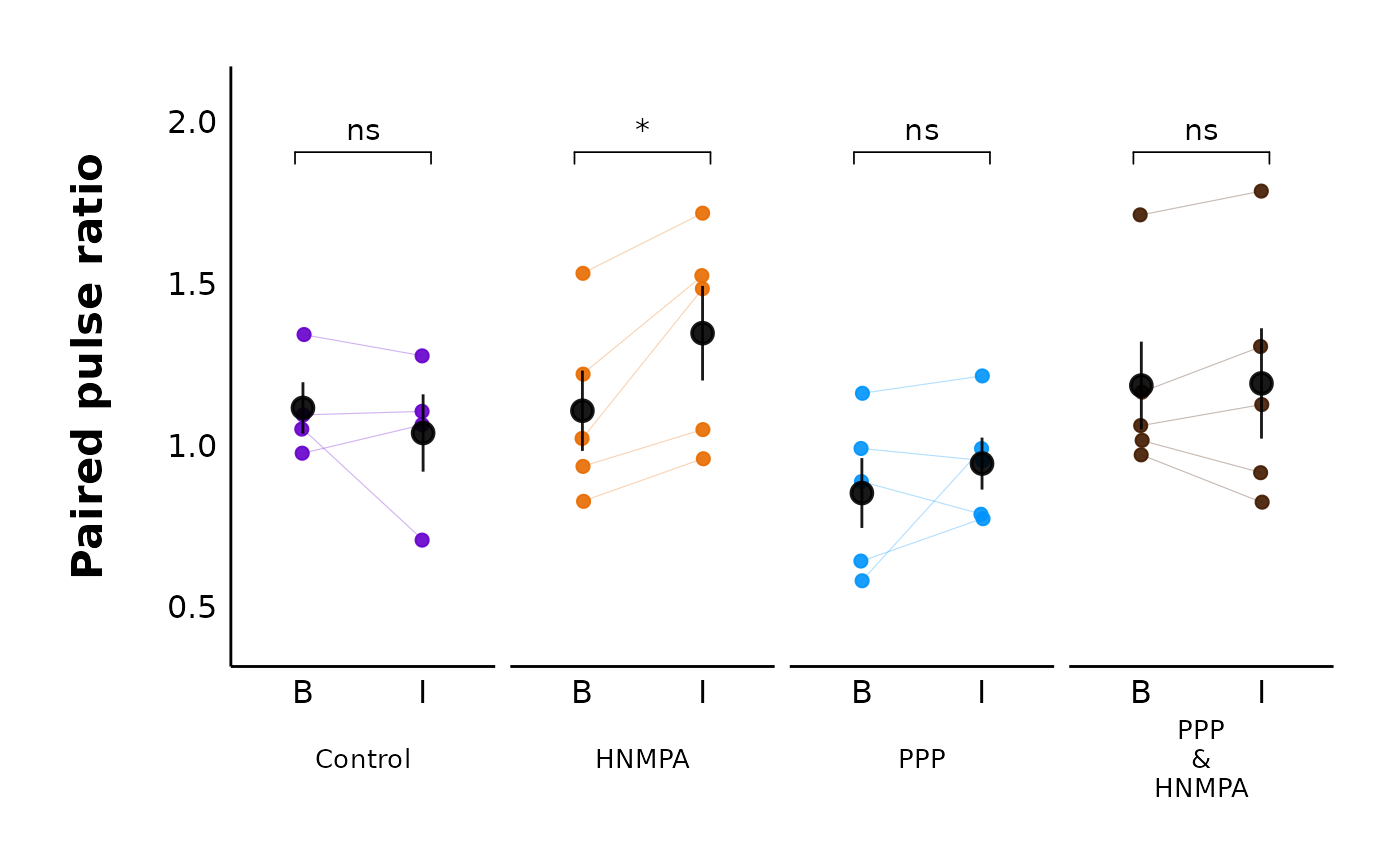
plot_PPR_data_multiple_treatments() creates a categorical scatter plot with
experimental state (i.e. grouped as baseline/before and after) and treatment
on the x-axis, and the paired-pulse ratio (PPR) on the y-axis. There are also
lines connecting the "before" data point to the "after" data point for each
letter. It is the same as plot_PPR_data_single_treatment() but for more
than one treatment.
Usage
plot_PPR_data_multiple_treatments(
data,
include_all_treatments = "yes",
list_of_treatments = NULL,
plot_category = 2,
included_sexes = "both",
male_label = "Male",
female_label = "Female",
baseline_label = "B",
post_hormone_label = "A",
test_type,
map_signif_level_values = F,
geom_signif_family = "",
geom_signif_text_size = 3,
geom_signif_size = 0.3,
treatment_colour_theme,
theme_options,
filename_suffix = "",
save_plot_png = "no",
ggplot_theme = patchclampplotteR_theme()
)Arguments
- data
Paired pulse ratio data generated from
make_PPR_data().- include_all_treatments
A character (
"yes"or"no") specifying if the plot will include data from all treatments. If"no", you must specify a list of treatments inlist_of_treatments.- list_of_treatments
A list of character values describing the treatments that will be in the plot. Defaults to
NULL, since include_all_treatments is"yes"by default.- plot_category
A numeric value specifying the category, which can be used to differentiate different protocol types. In the sample dataset for this package,
plot_category == 2represents experiments where insulin was applied continuously after a 5-minute baseline period.- included_sexes
A character value (
"both","male"or"female"). Useful if you want to have a plot with data from one sex only. Defaults to"both". If you choose a single sex, the resulting plot will have"-males-only"or"-females-only"in the file name.- male_label
A character value used to describe how males are encoded in the
sexcolumn of the dataframe used indata. This MUST match the value for male data in thesexcolumn, and it must be consistent across data sheets. Defaults to"Male".- female_label
A character value used to describe how females are encoded in the
sexcolumn of the dataframe used indata. This MUST match the value for female data in thesexcolumn, and it must be consistent across data sheets. This must be consistent in all data sheets. Defaults to"Female".- baseline_label
A character value for the x-axis label applied to the pre-hormone state. Defaults to
"Baseline".- post_hormone_label
A character value for x-axis label applied to the post-hormone or post-protocol state. Defaults to
"Post-hormone"but you will likely change this to the hormone or protocol name.- test_type
A character (must be
"wilcox.test","t.test"or"none") describing the statistical model used to create a significance bracket comparing the pre- and post-hormone groups.- map_signif_level_values
A
TRUE/FALSEvalue or a list of character values for mapping p-values. IfTRUE, p-values will be mapped with asterisks (e.g. \* for p < 0.05, for p < 0.01). IfFALSE, raw p-values will display. You can also insert a list of custom mappings or a function. For example, usemap_signif_level_values = function(p) if (p < 0.1) {round(p, 3)} else {"ns"}to only display the p-values when they are below 0.1.- geom_signif_family
A character value describing the font family used for the p-value annotations used by
ggsignif::geom_signif(). Defaults to""(empty value, will be replaced with default system font), but can be replaced with a named font. Use a package likeextrafontto load system fonts into R.- geom_signif_text_size
A numeric value describing the size of the text annotations (significance stars or p-values) on the plot. Defaults to
8.- geom_signif_size
A numeric value describing the size of the
geom_signifbracket size. Defaults to0.3, which is a good thickness for most applications.- treatment_colour_theme
A dataframe containing treatment names and their associated colours as hex values. See sample_treatment_names_and_colours for an example of what this dataframe should look like.
- theme_options
A dataframe containing theme options, defaults to
sample_theme_options. See sample_theme_options for an example of what this dataframe should look like and how you can customize these values.- filename_suffix
Optional character value to add a suffix to the filename of the .png file created with this plot. Could be useful if you have specified a custom list of treatments.
- save_plot_png
A character (
"yes"or"no"). If"yes", the plot will be saved as a .png usingggsave(). The filepath depends on the current type, but they will all go in subfolders belowFigures/in your project directory.- ggplot_theme
The name of a ggplot theme or your custom theme. This will be added as a layer to a ggplot object. The default is
patchclampplotteR_theme(), but other valid entries includetheme_bw(),theme_classic()or the name of a custom ggplot theme stored as an object.
Value
A ggplot object. If save_plot_png is defined as "yes", it will also
generate a .png file in the folder Figures/Evoked-currents/PPR relative
to the project directory.
Details
If you specify a test_type, the function will perform a paired t-test or
paired wilcox test and add brackets with significance stars through
ggsignif::geom_signif().
See also
plot_PPR_data_single_treatment() to plot changes in PPR for a single treatment. See make_PPR_data() for the function used to create the PPR data.
Examples
plot_PPR_data_multiple_treatments(
data = sample_PPR_df,
include_all_treatments = "yes",
plot_category = 2,
baseline_label = "B",
post_hormone_label = "I",
included_sexes = "both",
test_type = "t.test",
theme_options = sample_theme_options,
treatment_colour_theme = sample_treatment_names_and_colours
)