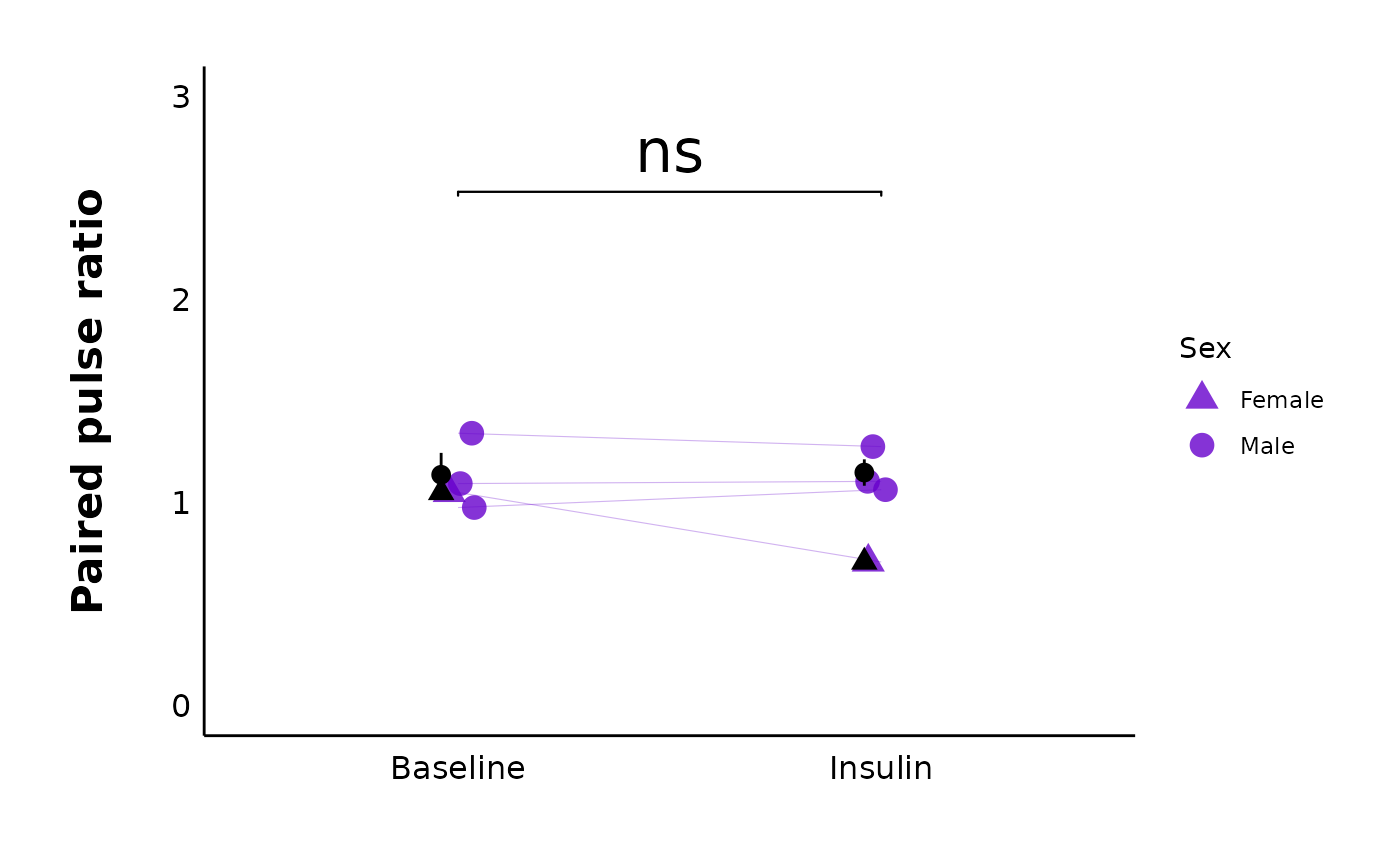
plot_PPR_data_one_treatment() creates a categorical scatter plot with
experimental state (i.e. baseline/before and after) on the x-axis and the
paired-pulse ratio (PPR) on the y-axis. There are also lines connecting the
"before" data point to the "after" data point for each letter.
Usage
plot_PPR_data_one_treatment(
data,
plot_treatment = "Control",
plot_category = 2,
baseline_label = "Baseline",
post_hormone_label = "Post-hormone",
test_type,
large_axis_text = "no",
geom_signif_size = 0.4,
treatment_colour_theme,
theme_options,
save_plot_png = "no",
ggplot_theme = patchclampplotteR_theme()
)Arguments
- data
Paired pulse ratio data generated from
make_PPR_data().- plot_treatment
A character value specifying the treatment you would like to plot (e.g. "Control").
plot_treatmentrepresents antagonists that were present on the brain slice, or the animals were fasted, etc.- plot_category
A numeric value specifying the category, which can be used to differentiate different protocol types. In the sample dataset for this package,
plot_category == 2represents experiments where insulin was applied continuously after a 5-minute baseline period.- baseline_label
A character value for the x-axis label applied to the pre-hormone state. Defaults to "Baseline".
- post_hormone_label
A character value for x-axis label applied to the post-hormone or post-protocol state. Defaults to "Post-hormone" but you will likely change this to the hormone or protocol name.
- test_type
A character (must be "wilcox.test", "t.test" or "none") describing the statistical model used to create a significance bracket comparing the pre- and post-hormone groups.
- large_axis_text
A character ("yes" or "no"). If "yes", a ggplot theme layer will be applied which increases the size of the axis text.
- geom_signif_size
A numeric value describing the size of the geom_signif bracket size. Defaults to 0.4, which is a good thickness for most applications.
- treatment_colour_theme
A dataframe containing treatment names and their associated colours as hex values. See sample_treatment_names_and_colours for an example of what this dataframe should look like.
- theme_options
A dataframe containing theme options. See sample_theme_options for an example of what this dataframe should look like.
- save_plot_png
A character ("yes" or "no"). If "yes", the plot will be saved as a .png using ggsave. The filepath depends on the current type, but they will all go in subfolders below Figures/ in your project directory.
- ggplot_theme
The name of a ggplot theme or your custom theme. This will be added as a layer to a ggplot object. The default is
patchclampplotteR_theme(), but other valid entries includetheme_bw(),theme_classic()or the name of a custom ggplot theme stored as an object.
Value
A ggplot object. If save_plot_png == "yes", it will also generate
a .png file in the folder Figures/Evoked-currents/PPR relative to the
project directory. The treatment will be included in the filename.
Details
If you specify a test_type, the function will perform a paired t-test or
paired wilcox test and add brackets with significance stars through
ggsignif::geom_signif().
See also
plot_PPR_data_multiple_treatments() to plot changes in PPR for multiple treatments.
Examples
plot_PPR_data_one_treatment(
data = sample_PPR_df,
plot_treatment = "Control",
plot_category = 2,
baseline_label = "Baseline",
post_hormone_label = "Insulin",
test_type = "t.test",
large_axis_text = "no",
treatment_colour_theme = sample_treatment_names_and_colours,
theme_options = sample_theme_options,
save_plot_png = "no"
)
#> Warning: Removed 2 rows containing missing values or values outside the scale range
#> (`geom_segment()`).